module vis::Graphs
Simple data visualization using graphs.
Usage
import vis::Graphs;
Dependencies
import lang::html::IO;
import lang::html::AST;
import util::IDEServices;
import Content;
import ValueIO;
Description
This modules provides a simple API to create graph visuals for Rascal (relational) data, based on Cytoscape.js.
This module is quite new and may undergo some tweaks in the coming time.
Benefits
- Easy to use for basic graph layouts.
function graph
A graph plot from a binary list relation.
Content graph(lrel[&T x, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler, str title="Graph", CytoLayout \layout=defaultCoseLayout(), CytoStyle nodeStyle=defaultNodeStyle(), CytoStyle edgeStyle=defaultEdgeStyle())
Examples
rascal>import vis::Graphs;
ok
rascal>graph([<x,x+1> | x <- [1..100]] + [<100,1>])
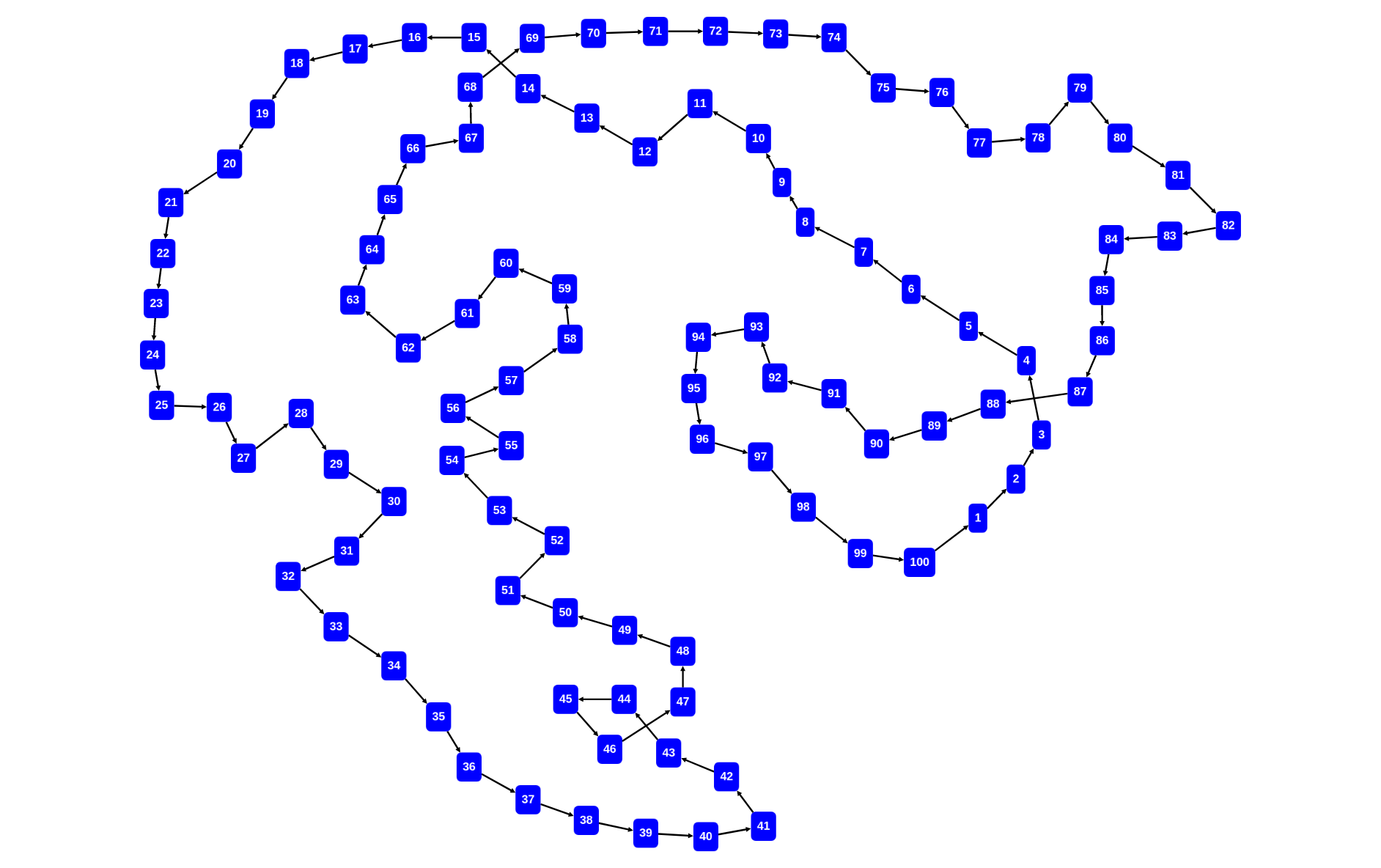
rascal>graph([<x,x+1> | x <- [1..100]] + [<100,1>], \layout=\defaultCircleLayout())
Providing locations as node identities automatically transforms them to node links:
rascal>import vis::Graphs;
ok
rascal>import IO;
ok
rascal>d = [<|std:///|, e> | e <- |std:///|.ls];
lrel[loc,loc]: [
<|std:///|,|std:///Boolean.rsc|>,
<|std:///|,|std:///Content.rsc|>,
<|std:///|,|std:///DateTime.rsc|>,
<|std:///|,|std:///Exception.rsc|>,
<|std:///|,|std:///Grammar.rsc|>,
<|std:///|,|std:///IO.rsc|>,
<|std:///|,|std:///List.rsc|>,
<|std:///|,|std:///ListRelation.rsc|>,
<|std:///|,|std:///Location.rsc|>,
<|std:///|,|std:///Map.rsc|>,
<|std:///|,|std:///Message.rsc|>,
<|std:///|,|std:///Node.rsc|>,
<|std:///|,|std:///ParseTree.rsc|>,
<|std:///|,|std:///Prelude$1.class|>,
<|std:///|,|std:///Prelude$2.class|>,
<|std:///|,|std:///Prelude$3.class|>,
<|std:///|,|std:///Prelude$4.class|>,
<|std:///|,|std:///Prelude$Backtrack.class|>,
<|std:///|,|std:///Prelude$ByteBufferBackedInputStream.class|>,
<|std:///|,|std:///Prelude$Distance.class|>,
<|std:///|,|std:///Prelude$Less.class|>,
<|std:///|,|std:///Prelude$NodeComparator.class|>,
<|std:///|,|std:///Prelude$ReleasableCallback.class|>,
<|std:///|,|std:///Prelude$Sorting.class|>,
<|std:///|,|std:///Prelude.class|>,
<|std:///|,|std:///Prelude.rsc|>,
<|std:///|,|std:///Relation.rsc|>,
<|std:///|,|std:///Set.rsc|>,
<|std:///|,|std:///String.rsc|>,
<|std:///|,|std:///Type.class|>,
<|std:///|,|std:///Type.rsc|>,
<|std:///|,|std:///ValueIO.rsc|>,
<|std:///|,|std:///analysis|>,
<|std:///|,|std:///index.md|>,
<|std:///|,|std:///lang|>,
<|std:///|,|std:///resource|>,
<|std:///|,|std:///util|>,
<|std:///|,|std:///vis|>
]
rascal>d += [<e,f> | <_, e> <- d, isDirectory(e), f <- e.ls];
lrel[loc,loc]: [
<|std:///|,|std:///Boolean.rsc|>,
<|std:///|,|std:///Content.rsc|>,
<|std:///|,|std:///DateTime.rsc|>,
<|std:///|,|std:///Exception.rsc|>,
<|std:///|,|std:///Grammar.rsc|>,
<|std:///|,|std:///IO.rsc|>,
<|std:///|,|std:///List.rsc|>,
<|std:///|,|std:///ListRelation.rsc|>,
<|std:///|,|std:///Location.rsc|>,
<|std:///|,|std:///Map.rsc|>,
<|std:///|,|std:///Message.rsc|>,
<|std:///|,|std:///Node.rsc|>,
<|std:///|,|std:///ParseTree.rsc|>,
<|std:///|,|std:///Prelude$1.class|>,
<|std:///|,|std:///Prelude$2.class|>,
<|std:///|,|std:///Prelude$3.class|>,
<|std:///|,|std:///Prelude$4.class|>,
<|std:///|,|std:///Prelude$Backtrack.class|>,
<|std:///|,|std:///Prelude$ByteBufferBackedInputStream.class|>,
<|std:///|,|std:///Prelude$Distance.class|>,
<|std:///|,|std:///Prelude$Less.class|>,
<|std:///|,|std:///Prelude$NodeComparator.class|>,
<|std:///|,|std:///Prelude$ReleasableCallback.class|>,
<|std:///|,|std:///Prelude$Sorting.class|>,
<|std:///|,|std:///Prelude.class|>,
<|std:///|,|std:///Prelude.rsc|>,
<|std:///|,|std:///Relation.rsc|>,
<|std:///|,|std:///Set.rsc|>,
<|std:///|,|std:///String.rsc|>,
<|std:///|,|std:///Type.class|>,
<|std:///|,|std:///Type.rsc|>,
<|std:///|,|std:///ValueIO.rsc|>,
<|std:///|,|std:///analysis|>,
<|std:///|,|std:///index.md|>,
<|std:///|,|std:///lang|>,
<|std:///|,|std:///resource|>,
<|std:///|,|std:///util|>,
<|std:///|,|std:///vis|>,
<|std:///analysis|,|std:///analysis/clustering|>,
<|std:///analysis|,|std:///analysis/diff|>,
<|std:///analysis|,|std:///analysis/flow|>,
<|std:///analysis|,|std:///analysis/formalconcepts|>,
<|std:///analysis|,|std:///analysis/grammars|>,
<|std:///analysis|,|std:///analysis/graphs|>,
<|std:///analysis|,|std:///analysis/linearprogramming|>,
<|std:///analysis|,|std:///analysis/m3|>,
<|std:///analysis|,|std:///analysis/statistics|>,
<|std:///analysis|,|std:///analysis/text|>,
<|std:///lang|,|std:///lang/aterm|>,
<|std:///lang|,|std:///lang/aut|>,
<|std:///lang|,|std:///lang/box|>,
<|std:///lang|,|std:///lang/c90|>,
<|std:///lang|,|std:///lang/csv|>,
<|std:///lang|,|std:///lang/diff|>,
<|std:///lang|,|std:///lang/dimacs|>,
<|std:///lang|,|std:///lang/dot|>,
<|std:///lang|,|std:///lang/html|>,
<|std:///lang|,|std:///lang/html5|>,
<|std:///lang|,|std:///lang/java|>,
<|std:///lang|,|std:///lang/javascript|>,
<|std:///lang|,|std:///lang/json|>,
<|std:///lang|,|std:///lang/manifest|>,
<|std:///lang|,|std:///lang/paths|>,
<|std:///lang|,|std:///lang/pico|>,
<|std:///lang|,|std:///lang/rascal|>,
<|std:///lang|,|std:///lang/rsf|>,
<|std:///lang|,|std:///lang/sdf2|>,
<|std:///lang|,|std:///lang/sexp|>,
<|std:///lang|,|std:///lang/smtlib2|>,
<|std:///lang|,|std:///lang/std|>,
<|std:///lang|,|std:///lang/uri|>,
<|std:///lang|,|std:///lang/xforms|>,
<|std:///lang|,|std:///lang/xml|>,
<|std:///lang|,|std:///lang/yaml|>,
<|std:///resource|,|std:///resource/jdbc|>,
<|std:///util|,|std:///util/Benchmark.class|>,
<|std:///util|,|std:///util/Benchmark.rsc|>,
<|std:///util|,|std:///util/Clipboard.class|>,
<|std:///util|,|std:///util/Clipboard.rsc|>,
<|std:///util|,|std:///util/Eval$EvaluatorInterruptTimer.class|>,
<|std:///util|,|std:///util/Eval$RascalRuntime.class|>,
<|std:///util|,|std:///util/Eval.class|>,
<|std:///util|,|std:///util/Eval.rsc|>,
<|std:///util|,|std:///util/FileSystem.rsc|>,
<|std:///util|,|std:///util/Highlight.rsc|>,
<|std:///util|,|std:///util/IDEServices.rsc|>,
<|std:///util|,|std:///util/IDEServicesLibrary.class|>,
<|std:///util|,|std:///util/Math.class|>,
<|std:///util|,|std:///util/Math.rsc|>,
<|std:///util|,|std:///util/Maybe.rsc|>,
<|std:///util|,|std:///util/Memo.rsc|>,
<|std:///util|,|std:///util/Monitor.class|>,
<|std:///util|,|std:///util/Monitor.rsc|>,
<|std:///util|,|std:///util/ObjectReader.class|>,
<|std:///util|,|std:///util/PathConfig$1.class|>,
<|std:///util|,|std:///util/PathConfig$LibResolverForMavenDependencies.class...
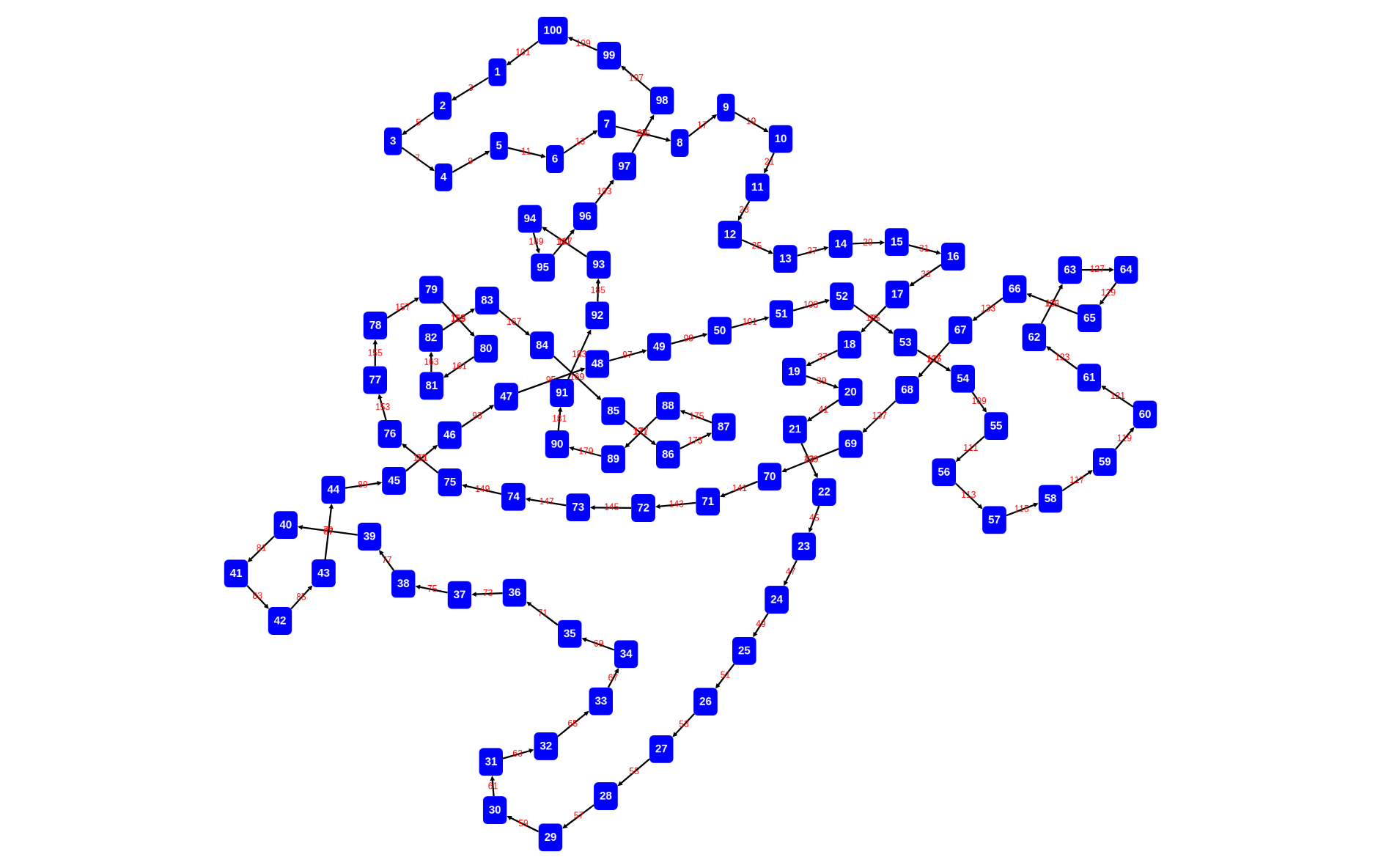
rascal>graph(d, \layout=defaultDagreLayout());

here we adapt the node labeler to show only the last file name in the path of the location:
rascal>graph(d, \layout=defaultDagreLayout(), nodeLabeler=str (loc l) { return l.file; });

function graph
A graph plot from a ternary list relation where the middle column is the edge label.
Content graph(lrel[&T x, &L edge, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, str title="Graph", CytoLayout \layout=defaultCoseLayout(), CytoStyle nodeStyle=defaultNodeStyle(), CytoStyle edgeStyle=defaultEdgeStyle())
Examples
rascal>import vis::Graphs;
ok
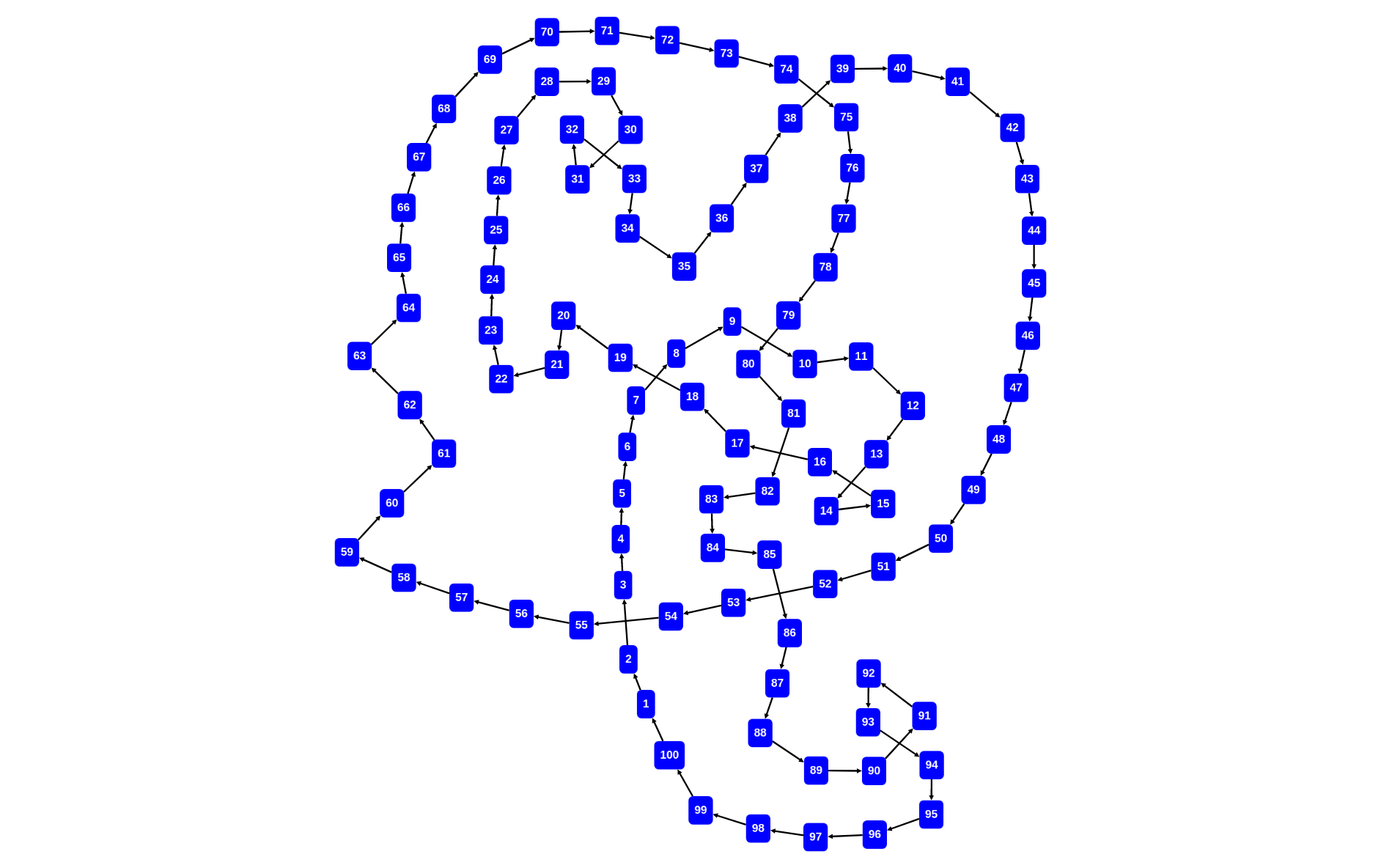
rascal>graph([<x,2*x+1,x+1> | x <- [1..100]] + [<100,101,1>])
function graph
A graph plot from a binary relation.
Content graph(rel[&T x, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler, str title="Graph", CytoLayout \layout=defaultCoseLayout(), CytoStyle nodeStyle=defaultNodeStyle(), CytoStyle edgeStyle=defaultEdgeStyle())
Examples
rascal>import vis::Graphs;
ok
rascal>graph({<x,x+1> | x <- [1..100]} + {<100,1>})
function graph
A graph plot from a ternary relation where the middle column is the edge label.
Content graph(rel[&T x, &L edge, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, str title="Graph", CytoLayout \layout=defaultCoseLayout(), CytoStyle nodeStyle=defaultNodeStyle(), CytoStyle edgeStyle=defaultEdgeStyle())
Examples
rascal>import vis::Graphs;
ok
rascal>graph({<x,2*x+1,x+1> | x <- [1..100]} + {<100,101,1>})
alias NodeLinker
loc (&T _id1)
function defaultNodeLinker
loc defaultNodeLinker(/loc l)
default loc defaultNodeLinker(&T _)
alias NodeLabeler
str (&T _id2)
function defaultNodeLabeler
str defaultNodeLabeler(/str s)
str defaultNodeLabeler(loc l)
default str defaultNodeLabeler(&T v)
alias EdgeLabeler
str (&T _source, &T _target)
function defaultEdgeLabeler
str defaultEdgeLabeler(&T _source, &T _target)
function cytoscape
Cytoscape cytoscape(list[CytoData] \data, \CytoLayout \layout=\defaultCoseLayout(), CytoStyle nodeStyle=defaultNodeStyle(), CytoStyle edgeStyle=defaultEdgeStyle())
function graphData
list[CytoData] graphData(rel[loc x, loc y] v, NodeLinker[loc] nodeLinker=defaultNodeLinker, NodeLabeler[loc] nodeLabeler=defaultNodeLabeler, EdgeLabeler[loc] edgeLabeler=defaultEdgeLabeler)
default list[CytoData] graphData(rel[&T x, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler)
list[CytoData] graphData(lrel[loc x, &L edge, loc y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler)
default list[CytoData] graphData(lrel[&T x, &L edge, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler)
list[CytoData] graphData(lrel[loc x, loc y] v, NodeLinker[loc] nodeLinker=defaultNodeLinker, NodeLabeler[loc] nodeLabeler=defaultNodeLabeler, EdgeLabeler[loc] edgeLabeler=defaultEdgeLabeler)
default list[CytoData] graphData(lrel[&T x, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler)
list[CytoData] graphData(rel[loc x, &L edge, loc y] v, NodeLinker[loc] nodeLinker=defaultNodeLinker, NodeLabeler[loc] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler)
default list[CytoData] graphData(rel[&T x, &L edge, &T y] v, NodeLinker[&T] nodeLinker=defaultNodeLinker, NodeLabeler[&T] nodeLabeler=defaultNodeLabeler, EdgeLabeler[&T] edgeLabeler=defaultEdgeLabeler)
data CytoNodeShape
data CytoNodeShape
= \ellipse()
| \triangle()
| \round-triangle()
| \rectangle()
| \round-rectangle()
| \bottom-round-rectangle()
| \cut-rectangle()
| \barrel()
| \rhomboid()
| \diamond()
| \round-diamond()
| \pentagon()
| \round-pentagon()
| \hexagon()
| \round-hexagon()
| \concave-hexagon()
| \heptagon()
| \round-heptagon()
| \octagon()
| \round-octagon()
| \star()
| \tag()
| \round-tag()
| \vee()
| \polygon()
;
data Cytoscape
data Cytoscape
= cytoscape(
list[CytoData] elements = [],
list[CytoStyleOf] style=[],
CytoLayout \layout = cytolayout()
)
;
data CytoData
data CytoData
= cytodata(CytoElement \data)
;
data CytoElement
data CytoElement
= \node(str id, str label=id, str editor="|none:///|")
| \edge(str source, str target, str id="<source>-<target>", str label="")
;
data CytoHorizontalAlign
data CytoHorizontalAlign
= left()
| center()
| right()
| auto()
;
data CytoVerticalAlign
data CytoVerticalAlign
= top()
| center()
| bottom()
| auto()
;
data CytoArrowHeadStyle
data CytoArrowHeadStyle
= triangle()
| \triangle-tee()
| \circle-triangle()
| \triangle-cross()
| \triangle-backcurve()
| vee()
| tee()
| square()
| circle()
| diamond()
| chevron()
| none()
;
data CytoTextWrap
data CytoTextWrap
= none()
| wrap()
| ellipses()
;
data CytoCurveStyle
data CytoCurveStyle
= bezier()
| \unbundled-bezier()
| straight()
| segments()
| \straight-triangle()
| taxi()
| haystack()
;
data CytoStyleOf
data CytoStyleOf
= cytoNodeStyleOf(
CytoSelector selector = \node(),
CytoStyle style = cytoNodeStyle()
)
| cytoEdgeStyleOf(
CytoSelector selector = \edge(),
CytoStyle style = cytoEdgeStyle()
)
;
function cytoNodeStyleOf
CytoStyleOf cytoNodeStyleOf(CytoStyle style)
function cytoEdgeStyleOf
CytoStyleOf cytoEdgeStyleOf(CytoStyle style)
function defaultNodeStyle
CytoStyle defaultNodeStyle()
function defaultEdgeStyle
CytoStyle defaultEdgeStyle()
data CytoFontWeight
data CytoFontWeight
= normal()
| lighter()
| bold()
| bolder()
;
data CytoStyle
data CytoStyle
= cytoNodeStyle(
str width = "label",
str padding = "10pt",
str color = "white",
str \text-opacity = "100%",
str \font-family = "",
str \font-size = "12pt",
str \font-style = "",
CytoFontWeight \font-weight = normal(),
str \background-color = "blue",
str label = "data(label)",
CytoNodeShape shape = CytoNodeShape::ellipse(),
CytoHorizontalAlign \text-halign = CytoHorizontalAlign::center(),
CytoVerticalAlign \text-valign = CytoVerticalAlign::\top(),
CytoTextWrap \text-wrap = CytoTextWrap::none(),
str \text-max-width = "100px",
CytoHorizontalAlign \text-justification = CytoHorizontalAlign::center(),
int \line-height = 1
)
| cytoEdgeStyle(
int width = 3,
str \line-color = "black",
str color = "red",
str \target-arrow-color = "black",
str \source-arrow-color = "black",
CytoArrowHeadStyle \target-arrow-shape = CytoArrowHeadStyle::triangle(),
CytoArrowHeadStyle \source-arrow-shape = CytoArrowHeadStyle::none(),
CytoCurveStyle \curve-style = CytoCurveStyle::bezier(),
int \source-text-offset = 1,
int \target-text-offset = 1,
str label = "data(label)"
)
;
data CytoSelector
data CytoSelector
= \node()
| \edge()
;
data CytoLayoutName
data CytoLayoutName
= grid()
| circle()
| breadthfirst()
| cose()
| dagre()
;
function hierarchical
An alias for dagre layout for documentation purposes.
CytoLayoutName hierarchical()
Dagre is a hierarchical graph layout.
data CytoLayout
data CytoLayout (CytoLayoutName name = dagre(), bool animate=false)
= cytolayout()
| breadthfirstLayout(
CytoLayoutName name = CytoLayoutName::breadthfirst(),
num spacingFactor= 1,
list[str] roots = [],
bool circle=false,
bool grid=!circle,
bool directed=false
)
| gridLayout(
CytoLayoutName name = CytoLayoutName::grid(),
int rows=2,
int cols=2,
bool avoidOverlap=true,
num spacingFactor=.1
)
| circleLayout(
CytoLayoutName name = CytoLayoutName::circle(),
bool avoidOverlap=true,
num spacingFactor=.1
)
| coseLayout(
CytoLayoutName name = cose()
)
| dagreLayout(
CytoLayoutName name = dagre(),
num spacingFactor = .1
)
;
function defaultCoseLayout
CytoLayout defaultCoseLayout()
function defaultCircleLayout
CytoLayout defaultCircleLayout(bool avoidOverlap=true, num spacingFactor=.1)
function defaultGridLayout
CytoLayout defaultGridLayout(int rows=2, int cols=rows, bool avoidOverlap=true, num spacingFactor=.1)
function defaultBreadthfirstLayout
CytoLayout defaultBreadthfirstLayout(num spacingFactor=.1, bool circle=false, bool grid=!circle, bool directed=false)
function defaultDagreLayout
CytoLayout defaultDagreLayout(num spacingFactor=1)
function graphServer
This is the main server generator for any graph value.
Response (Request) graphServer(Cytoscape ch)
Given a Graph value this server captures the value and serves it as a JSON value to the HTML client generated by plotHTML.
function plotHTML
Default HTML wrapper for a chart.
HTMLElement plotHTML()